Lecture 6: Pacific Biosciences Sequencing Technology
Lecture 6: Pacific Biosciences Sequencing Technology
Guest Lecture by Stephen Turner, CTO and co-founder of PacBio
Wednesday 13 April 2016
Scribed by the course staff
Topics
Technology background
A brief overview of the PacBio’s single molecule real time (SMRT) sequencing technology can be found here.
During his PhD at Cornell, Stephen Turner and his collaborators used near-field scanning optical microscopy (NSOM) to observe what was happening within a microscopic well. The method involved taking a waveguide, heating it up and pulling it to a fine point, and drilling a subwavelength aperture. Turner and colleagues were able to see volumes as small as \(20*10^{-21}\) liters, resulting in the invention of the zero mode waveguide (ZMW).
By attaching a polymerase to the bottom of the well and carefully tagging the dNTPs, one can observe a sequence of colored bursts of light from the nucleotides as they get incorporated. The polymerase is located where the electric field is expanding and the illumination was greatest.
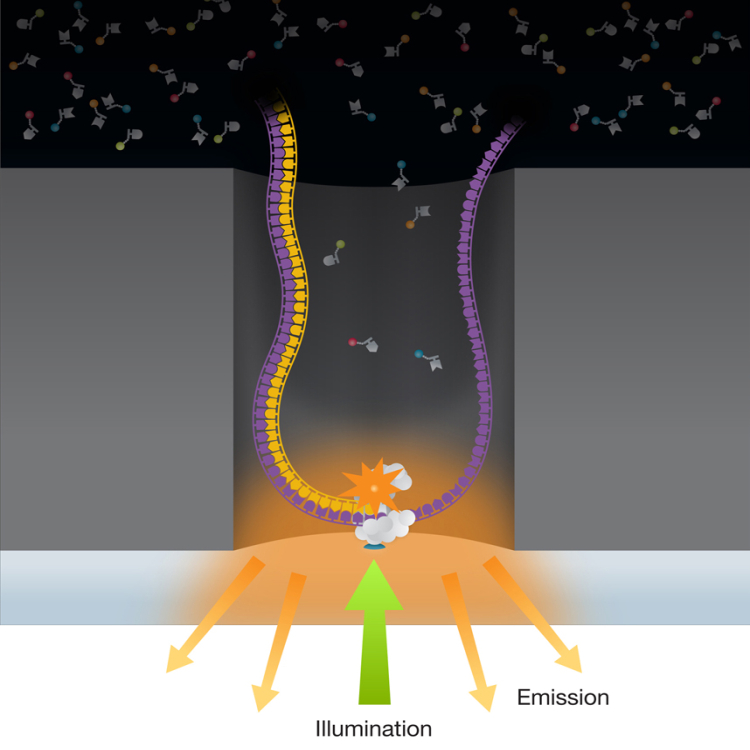
Compared to Illumina, this system is more sensitive optically, allowing for real time sequencing. With the ZMW technology, once the pyrophosphate is generated during incorporation of the base, it goes away forever. During the polymerization, there are no moving parts other than the polymerase. Because Illumina halts synthesis, the technology suffers from phasing issues. For example, the polymerase may not start up again.
While the nucleotides are floating in the ZMW, they generate very short baseline diffusion signals. When a base is integrated, the pulse of light generated is about 10000 times as long as the baseline diffusion signals. The timings between pulses (corresponding to bases) are not regular, however, due to variable kinetics as a function of local DNA chemistry. This allows one to see chemical modifications of DNA without any preparation ahead of time.
As a sequencing machine, DNA polymerase has impressive performance parameters. Not only can it sequence almost error free at a rate of 750 bp/s, but the read length can be on the order of millions. A polymerase can sequence an entire genome with 12 picograms of material. PacBio’s technology can sequence at about 3 bp/s, and their machine has 150000 ZMWs sequencing in parallel. The read length is currently over 10000 and has been doubling with every polymerase release.
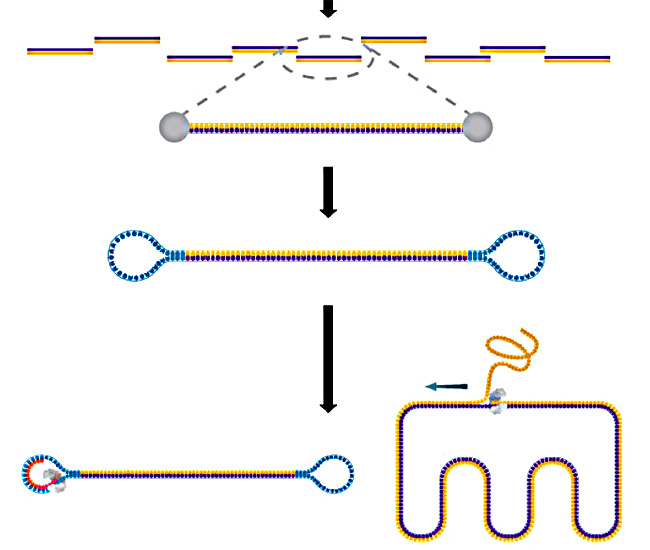
A unique feature of the PacBio technology is how it uses genomic DNA. Like other technologies, PacBio uses universal adapters, but unlike other technologies, the adapters are hairpin adapters. The polymerase loops around during synthesis, displacing strands they encounter.
Error rates
At the single molecule level, PacBio has about a 10% error rate, but they suspect this rate will decrease with appropriately constructed algorithms. In a single molecule, the errors are random. Other technologies such as Oxford Nanopore suffer from systematic biases that cannot be corrected even with infinite reads. With 50-fold coverage, PacBio reads can do more confident calls than any other technology. Large repeats still pose a significant challenge, however.
Errors are caused mainly due to
- A nucleotide becoming incorporated unusually quickly. Since the time between nucleotide incorporations is exponentially distributed, the most frequent incorporation times are close to 0 (hard to sample).
- The system making the wrong call, which happens about 1% of the time.
Long read advantages
One of the main advantages of long reads is that one can sequence the entire genome of some organisms. Because PacBio does not involve PCR (which cannot amplify certain sequences such as palindromic sequences), they can sequence areas of genomes that are challenging to other technologies. Additionally, PacBio is an asynchronous system and does not involve any washing steps, resulting in less bias. This unbiasedness remains even as GC content is varied. With short reads, the coverage may be quite nonuniform due to GC content.
A recent PacBio success story involves the sequencing of the HIV genome. With short reads, tracking the evolution of a particular case of HIV is laborious. HIV is constantly adapting to escape the patient’s immune system, but PacBio’s long reads allowed scientists to follow this evolution.
PacBio also allowed scientists to find unexpected presence of genetic material within an organism. For example, PacBio long reads allowed scientists to find a sequence corresponding to Shiga toxin within the genome of a strain of E. coli. More details can be found here.